DynDom3D - Protein Domain Motion Analysis
DynDom3D is a new program for the analysis of domain movements in large, multi-chain, biomolecular complexes. Use the form below to run the program or click on the "About DynDom3D" link to find out more about DynDom3D and how to interpret results.
Please note: An alignment is applied to the PDB files prior to processing by DynDom3D. The alignment matches models and the chains within those models, removing residues and atoms as necessary in order to achieve one-to-one correspondence which is required by DynDom3D. An example alignment can be seen:
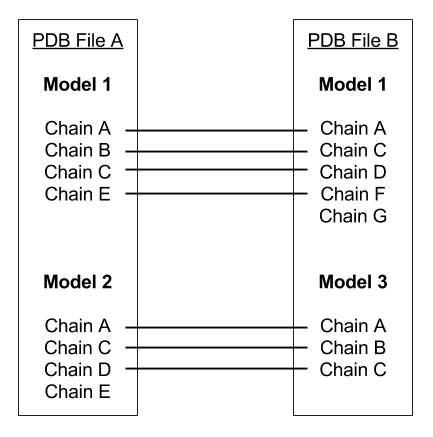 In this example, the residues under Chain E in Model 2 of PDB File A and, Chain G in Model 1 of PDB File B would not be present in the final PDB file.
In this example, the residues under Chain E in Model 2 of PDB File A and, Chain G in Model 1 of PDB File B would not be present in the final PDB file.