5'-Nucleotidase
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
GLN-353
ASN-354
1.6
1.5
-10.9
22.7
101.5
98.0
-0.4
ASN-354
LYS-355
2.7
2.5
-37.8
7.5
114.7
120.9
-14.8
LYS-355
GLY-356
5.5
5.3
-179.5
-113.6
158.7
168.7
154.0
GLY-356
LYS-357
6.8
3.8
-70.7
-7.0
110.2
121.3
-78.7
LYS-357
ALA-358
4.7
2.7
-10.5
-5.5
123.8
102.0
-15.6
ALA-358
GLN-359
8.3
6.5
6.8
2.3
14.5
44.8
17.3
GLN-359
LEU-360
8.0
7.3
-7.8
-9.3
120.7
156.7
-33.2
LEU-360
GLU-361
4.4
4.9
35.1
10.0
76.0
67.0
56.7
GLU-361
VAL-362
6.5
7.0
-13.1
6.9
116.8
117.6
-24.1
VAL-362
LYS-363
9.4
9.8
12.0
-8.5
21.4
6.1
25.9
LYS-363
ILE-364
10.6
10.9
3.3
0.7
48.4
60.5
7.6
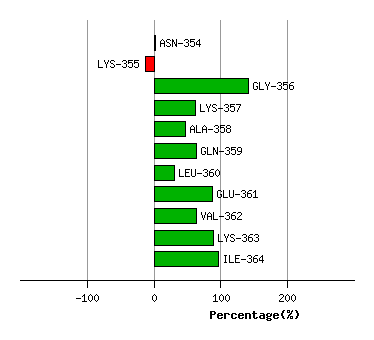
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees