Pyruvate Kinase
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
GLY-86
PRO-87
4.7
4.4
7.3
6.3
78.8
94.7
-5.8
PRO-87
GLU-88
1.0
0.7
-72.6
3.8
38.9
43.1
133.3
GLU-88
ILE-89
3.6
3.5
7.3
1.7
86.3
74.5
-24.1
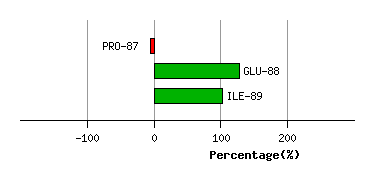
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
LEU-186
PRO-187
5.6
4.1
-13.4
-8.7
65.7
70.2
11.5
PRO-187
ALA-188
5.0
3.5
-16.4
10.2
117.3
111.1
-14.9
ALA-188
VAL-189
6.2
4.9
7.2
-2.4
102.7
118.6
-5.9

Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees