Vitamin D Hydroxylase
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
PRO-146
VAL-147
6.7
6.1
1.5
6.6
131.8
130.6
-5.3
VAL-147
GLN-148
9.0
9.1
-7.7
-0.7
110.8
102.3
1.9
GLN-148
VAL-149
9.7
9.7
0.1
-0.7
93.1
104.2
-1.1
VAL-149
ILE-150
6.2
6.6
-1.5
-6.9
19.2
8.9
36.2
ILE-150
CYS-151
6.2
6.4
4.9
-7.4
100.2
110.9
1.3
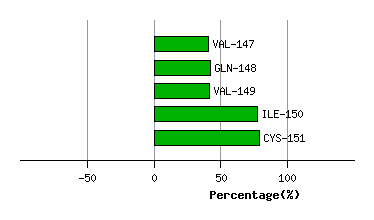
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
LEU-211
ALA-212
14.1
13.1
-9.0
8.1
98.8
110.7
26.9
ALA-212
VAL-213
16.5
15.9
-7.5
-15.2
119.5
120.8
-40.2
VAL-213
SER-214
14.5
14.1
-174.3
102.3
159.1
159.6
-473.6
SER-214
ASP-215
16.6
12.3
-152.4
17.1
88.2
127.1
157.1
ASP-215
GLU-216
17.7
15.8
145.3
-9.3
56.7
69.7
541.7
GLU-216
ASP-217
21.2
17.3
-6.2
-76.7
166.0
131.9
-445.5
ASP-217
GLY-218
19.8
14.8
76.3
-153.8
90.1
81.8
-63.1
GLY-218
ASP-219
18.7
11.7
-10.0
-29.5
84.9
109.8
-0.9
ASP-219
ARG-220
15.0
10.7
-175.0
88.4
36.3
35.0
554.0
ARG-220
LEU-221
13.0
10.8
152.1
-72.8
79.0
112.4
-59.0
LEU-221
SER-222
9.7
8.5
-17.3
-16.1
121.2
124.5
-71.8
SER-222
GLN-223
9.8
7.7
-0.8
-6.8
54.3
56.2
35.0
GLN-223
GLU-224
10.9
9.2
0.0
0.6
83.9
77.8
6.8
GLU-224
GLU-225
7.9
6.7
1.7
1.0
129.0
133.1
-31.6
GLU-225
LEU-226
5.6
4.0
-1.4
2.4
16.5
25.3
14.3
LEU-226
VAL-227
7.7
6.8
1.5
0.1
114.1
115.5
-19.2
VAL-227
ALA-228
8.1
7.7
2.1
-7.2
94.8
107.6
6.9
ALA-228
MET-229
4.8
4.8
2.9
3.7
144.4
146.5
-21.0
MET-229
ALA-230
3.2
3.1
-1.9
2.9
32.1
38.2
1.3
ALA-230
MET-231
5.7
6.2
2.2
-1.5
98.8
102.7
13.1
MET-231
LEU-232
6.4
7.0
-3.2
-2.4
74.6
60.9
16.5
LEU-232
LEU-233
3.5
4.4
0.7
-5.7
169.0
164.4
38.5
LEU-233
LEU-234
2.5
3.3
4.6
-4.6
124.3
124.8
-5.5
LEU-234
ILE-235
6.1
6.9
4.1
1.8
89.1
104.8
-12.9
ILE-235
ALA-236
7.4
8.1
-2.2
17.6
64.1
48.6
-67.0
ALA-236
GLY-237
5.7
5.7
-14.2
35.5
5.8
25.2
-103.0
GLY-237
HIS-238
4.6
5.6
-11.3
-11.7
51.7
64.0
45.4
HIS-238
GLU-239
7.8
9.4
50.7
-65.8
110.1
100.7
51.6
HIS-238
GLU-239
7.8
9.4
50.7
-65.8
110.1
100.7
51.6

Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
HIS-238
GLU-239
7.8
9.4
50.7
-65.8
110.1
100.7
51.6
HIS-238
GLU-239
7.8
9.4
50.7
-65.8
110.1
100.7
51.6
GLU-239
THR-240
10.8
10.5
49.9
-65.9
114.9
108.5
60.8
THR-240
THR-241
9.6
9.0
-20.7
12.0
15.8
8.0
27.6

Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees