B-Raf Proto-Oncogene Serine/threonine-Protein Kinase
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
LYS-482
MET-483
8.5
8.8
-14.7
40.7
119.7
123.8
45.6
MET-483
LEU-484
6.0
6.6
-28.0
-6.8
127.5
106.9
-52.8
LEU-484
ASN-485
5.4
5.6
-164.6
-152.7
156.4
168.8
122.5
ASN-485
VAL-486
5.6
7.0
105.8
-85.1
44.9
77.1
36.8
VAL-486
THR-487
4.4
4.2
26.1
-39.4
89.0
58.9
-34.7

Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
VAL-503
LEU-504
5.1
4.8
-0.5
4.2
108.7
97.0
5.7
LEU-504
ARG-505
4.0
3.4
5.9
-22.5
55.4
49.4
-33.4
ARG-505
LYS-506
1.0
0.9
29.7
-18.3
21.5
18.9
15.1
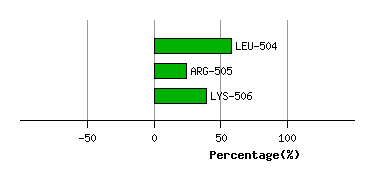
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees