Bifunctional P-450/nadph-P450 Reductase
(All numbering and residues are taken from first PDB file)
![]()
![]()
Bending Residue Dihedral Analysis
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
TYR-166
ARG-167
15.6
15.2
7.7
-12.1
42.1
43.7
10.8
ARG-167
ASP-168
13.1
12.7
13.0
2.6
35.0
30.4
-40.4
ASP-168
GLN-169
13.1
11.4
-35.7
22.2
107.1
83.9
-42.2
GLN-169
PRO-170
9.3
8.6
-39.3
-3.9
32.8
16.1
185.1
PRO-170
HIS-171
6.6
6.7
8.8
9.5
131.5
124.3
-28.6
HIS-171
PRO-172
6.2
6.6
-5.8
3.8
106.1
104.9
-11.3
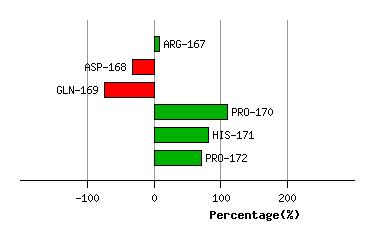
Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees
Residue
iResidue
i+1Distance of hinge axis to residue i in
(A) Distance of hinge axis to residue i in
(A) Change in
(deg) Change in
(deg) Angle of psi(i) axis to hinge axis
(deg) Angle of psi(i) axis to hinge axis
(deg) Percentage Progress
HIS-236
MET-237
6.9
6.7
-15.4
-10.0
70.2
92.6
-6.9
MET-237
LEU-238
6.1
5.9
6.7
22.4
51.3
53.0
32.6
LEU-238
ASN-239
3.9
3.2
-7.0
21.1
167.5
160.3
64.0

Graph shows rotational transition at bending residues and can be used
to identify hinge bending residues.
Probably only informative for interdomain rotations greater than 20 degrees