
Gtp-Binding Protein Lepa
(All numbering and residues are taken from first PDB file)
![]()
![]()
| Run Details | Domains | Sequence | Morph | Domain Pairs | Contact Graph |
DynDom run details
| Property | Value |
|---|---|
| Name | Gtp-Binding Protein Lepa |
| Conformer 1 (PDB) |
3cb4 (A) |
| Conformer 2 (PDB) |
3cb4 (C) |
| Window Length | 5 |
| Minimum ratio | 1.0 |
| Minimum domain size | 20 |
Domains
| Domain | Size | Backbone RMSD (A) |
Residues |
|---|---|---|---|
| 1 | 261 | 0.51 |
|
| 2 | 260 | 0.44 |
|
Sequence
|
Morph
This morph was created using the MorphIt_Pro protein morphing technique.
|
Domain Pairs
| Property | Value |
|---|---|
|
Fixed Domain
( blue ) |
1 |
|
Moving Domain
( red ) |
2 |
|
Rotation Angle
(deg) |
4.8 |
|
Translation
(A) |
0.1 |
|
Closure
(%) |
99.8 |
|
Bending Residues
( green ) |
|
| Bending Region Analysis | |
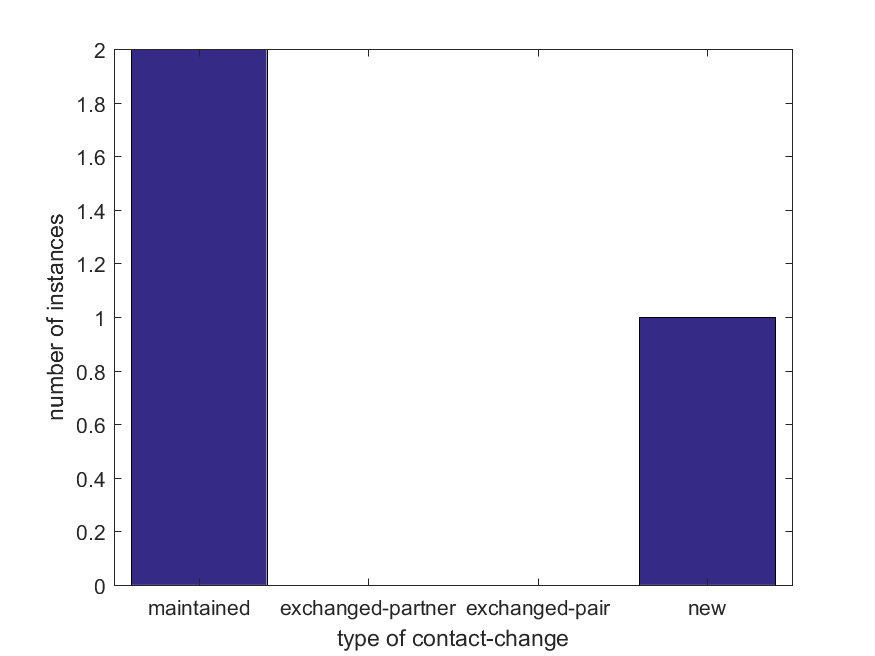
Dynamic Contact Graph
Full-size image
×Conformer 1 Contact:
Residue 1 ———› Residue 2
Residue 1 ———› Residue 2
Conformer 2 Contact:
Residue 2
———› Residue 1
Residue 2 ———› Residue 1
Movement classification: Mixed

PyMOL Script
|
