DynDom Description |
DynDom is a program that determines protein domains, hinge axes and amino acid residues involved in the hinge bending. It is fully automated.
You can use DynDom if you have two conformations of the same protein. These may be two X-ray structures, or structures generated using simulation techniques such as molecular dynamics or normal mode analysis.
The application of DynDom provides a view of the conformational change that is easily understood. The conformational change may be quite complicated in detail, but by using DynDom you can visualize it as involving the movement of domains as quasi-rigid bodies. The analysis of a conformational change in terms of domain movements only makes sense if the interdomain deformation is at least comparable to the intradomain deformation. You can use DynDom to assess this, but the results could be misleading if this is not the case.
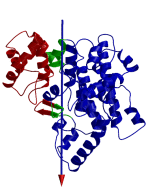
DynDom allows you to visualize the domain motion in terms of the rotation of one domain relative to another. Here we see the motion of the red domain relative to the blue domain that occurs upon substrate binding in citrate synthase. The arrow indicates the hinge axis and green those regions where the hinge bending takes place.
|
For further details email : sjh@cmp.uea.ac.uk |
DynDom © University of East Anglia 2015. All Rights Reserved. |
|